- Overview
- Summary
- Learning Objectives
- Missing Data
- Identifying Missing Data
- Recode Missing Data
- Excluding Missing Data
- Basic Missing Value Imputation Techniques
- Replace the missing value(s) with some constant, specified by the analyst
- Replace the missing value(s) with the mean, median or mode
- More Complex Approaches to Missing Value Imputation
- Special values
- Identifying Special Values
- Remark: Useful
applyfamily functions
- Optional Reading: Checking for Obvious Inconsistencies or Errors
- Optional Reading: Correction of Obvious Inconsistencies or Errors
- Additional Resources and Further Reading
- References
Module 5
Scan: Missing Values
Overview
Summary
Dealing missing values is an unavoidable task in the data
preprocessing. For almost every data set, we will encounter some missing
values. So, it is important to know how R handles missing values and how
they are represented. In this module, first you will learn how the
missing values and special values are represented in R. Then, you will
learn how to identify, recode and exclude missing values. Moreover, we
will cover missing value imputation techniques briefly. Note that the
missing value analysis and the missing value imputation are broader
concepts that would be a standalone topic of another course. Interested
readers may refer to the books and resources in the additional
resources and further reading section for further
details.
The analysts may also need to check and repair inconsistencies and
missing values in data records by using information from valid values
and validation rules restricting the data.
In this module, I also briefly introduced the deductive
package, and useful functions to correct the obvious errors and
inconsistencies in a given data set. However, this part of module left
as an optional reading for students.
Learning Objectives

The learning objectives of this module are as follows:
- Learn how missing and special values are represented in the data set.
- Identify missing values in the data set.
- Learn how to recode missing values.
- Learn the functions for removing missing values.
- Learn commonly used approaches to impute/replace missing value(s).
- (Optional) Check and correct obvious inconsistencies and errors in the data set.
Missing Data
In R, a numeric missing value is represented by NA (NA
stands for “not available”), while character missing values are
represented by <NA>. In addition to NA
and <NA>, some other values may represent missing
values (i.e. 99, ., .., just
space, or NULL) depending on the software (i.e., Excel,
SPSS etc.) that you import in your data.
Let’s have a look at the pet1.csv data:
library(readr)
pet1 <- read_csv("data/pet1.csv")## Rows: 30 Columns: 6
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (4): State, Region, Animal_Name, Colour_primary
## dbl (2): id, Reference
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.head(pet1)## # A tibble: 6 × 6
## id State Region Reference Animal_Name Colour_primary
## <dbl> <chr> <chr> <dbl> <chr> <chr>
## 1 118269 Victoria Ballarat NA Jack Wilson Brown
## 2 106347 Victoria Ballarat NA Eva Black And White
## 3 156347 Victoria Wyndham NA <NA> TRI
## 4 63947 Victoria Geelong NA Archie White/Brown
## 5 79724 Victoria Ballarat NA Susie Brown
## 6 43442 Victoria Geelong NA Pearl Tri ColourNote that, as we read this data from a .csv file, missing values are
represented as NA for the integer reference
variable where else <NA> for the character
Animal_Name variable.
However, let’s look at another example SPSS data file named population_NA.sav:
library(foreign)
population_NA <- read.spss("data/population_NA.sav", to.data.frame = TRUE)
population_NA## Region X.2013 X.2014 X.2015 X.2016
## 1 ISL 3.21 3.25 3.28 3.32
## 2 CAN 3.87 3.91 3.94 3.99
## 3 RUS 7.83 7.85 7.87 ..
## 4 COL 41.27 41.74 NA ..
## 5 ZAF 43.53 44.22 NA ..
## 6 LTU 47.42 46.96 46.63 46.11
## 7 MEX 60.43 61.10 61.76 62.41
## 8 IND 394.85 NA NA ..
## 9 NLD 497.64 499.59 501.68 504.01
## 10 KOR 504.92 506.97 508.91 510.77As you see in the data frame, there are two different representations
for the missing values: one is NA, the other is
.. . Therefore, we need to be careful about different
representations of the missing values while importing the data from
other software.
Identifying Missing Data
To identify missing values we will use is.na() function
which returns a logical vector with TRUE in the element
locations that contain missing values represented by NA.
is.na() will work on vectors, lists, matrices, and data
frames.
Here are some examples of is.na() function:
# create a vector with missing data
x <- c(1:4, NA, 6:7, NA)
x## [1] 1 2 3 4 NA 6 7 NAis.na(x)## [1] FALSE FALSE FALSE FALSE TRUE FALSE FALSE TRUE# create a data frame with missing data
df <- data.frame(col1 = c(1:3, NA),
col2 = c("this", NA,"is", "text"),
col3 = c(TRUE, FALSE, TRUE, TRUE),
col4 = c(2.5, 4.2, 3.2, NA))
# identify NAs in full data frame
is.na(df)## col1 col2 col3 col4
## [1,] FALSE FALSE FALSE FALSE
## [2,] FALSE TRUE FALSE FALSE
## [3,] FALSE FALSE FALSE FALSE
## [4,] TRUE FALSE FALSE TRUE# identify NAs in specific data frame column
is.na(df$col4)## [1] FALSE FALSE FALSE TRUETo identify the location or the number of
NAs we can use the which() and sum()
functions:
# identify location of NAs in vector
which(is.na(x))## [1] 5 8# identify count of NAs in data frame
sum(is.na(df))## [1] 3More convenient way to compute the total missing values in
each column is to use colSums():
colSums(is.na(df))## col1 col2 col3 col4
## 1 1 0 1Recode Missing Data
We can use normal subsetting and assignment operations in order to
recode missing values; or recode specific indicators that represent
missing values.
For instance, we can recode missing values in vector x
with the mean values in x. To do this, first we need to
subset the vector to identify NAs and then assign these
elements a value. Here is an example:
# create vector with missing data
x <- c(1:4, NA, 6:7, NA)
x## [1] 1 2 3 4 NA 6 7 NA# recode missing values with the mean (also see "Missing Value Imputation Techniques" section)
x[is.na(x)] <- mean(x, na.rm = TRUE)
x## [1] 1.000000 2.000000 3.000000 4.000000 3.833333 6.000000 7.000000 3.833333Similarly, if missing values are represented by another value
(i.e. ..) we can simply subset the data for the elements
that contain that value and then assign a desired value to those
elements.
Remember that population_NA data frame has missing
values represented by ".." in the X.2016
column. Now let’s change ".." values to
NA’s.
# population_NA data frame has missing values represented by ".." in the X.2016 column.
population_NA$X.2016## [1] "3.32 " "3.99 " ".. " ".. " ".. " "46.11 " "62.41 "
## [8] ".. " "504.01 " "510.77 "# Note the white spaces after ..'s and change ".. " values to NAs
population_NA[population_NA == ".. " ] <- NA
population_NA## Region X.2013 X.2014 X.2015 X.2016
## 1 ISL 3.21 3.25 3.28 3.32
## 2 CAN 3.87 3.91 3.94 3.99
## 3 RUS 7.83 7.85 7.87 <NA>
## 4 COL 41.27 41.74 NA <NA>
## 5 ZAF 43.53 44.22 NA <NA>
## 6 LTU 47.42 46.96 46.63 46.11
## 7 MEX 60.43 61.10 61.76 62.41
## 8 IND 394.85 NA NA <NA>
## 9 NLD 497.64 499.59 501.68 504.01
## 10 KOR 504.92 506.97 508.91 510.77If we want to recode missing values in a single data frame variable,
we can subset for the missing value in that specific variable of
interest and then assign it the replacement value. For example, in the
following example, we will recode the missing value in col4
with the mean value of col4.
# data frame with missing data
df <- data.frame(col1 = c(1:3, NA),
col2 = c("this", NA,"is", "text"),
col3 = c(TRUE, FALSE, TRUE, TRUE),
col4 = c(2.5, 4.2, 3.2, NA))
# recode the missing value in col4 with the mean value of col4
df$col4[is.na(df$col4)] <- mean(df$col4, na.rm = TRUE)
df## col1 col2 col3 col4
## 1 1 this TRUE 2.5
## 2 2 <NA> FALSE 4.2
## 3 3 is TRUE 3.2
## 4 NA text TRUE 3.3Note that, replace_na() function from tidyr package can
also be used to replace NA values. For more information and
examples see here.
Excluding Missing Data
A common method of handling missing values is simply to omit the
records or fields with missing values from the analysis. However, this
may be dangerous, since the pattern of missing values may in fact be
systematic, and simply deleting records with missing values would lead
to a biased subset of the data.
Some authors recommend that if the amount of missing data is very
small relatively to the size of the data set (up to 5%), then leaving
out the few values with missing features would be the best strategy in
order not to bias the analysis. When this is the case, we can exclude
missing values in a couple different ways.
If we want to exclude missing values from mathematical operations, we
can use the na.rm = TRUE argument. If you do not exclude
these values, most functions will return an NA. Here are
some examples:
# create a vector with missing values
x <- c(1:4, NA, 6:7, NA)
# including NA values will produce an NA output when used with mathematical operations
mean(x)## [1] NA# excluding NA values will calculate the mathematical operation for all non-missing values
mean(x, na.rm = TRUE)## [1] 3.833333We may also want to subset our data to obtain complete observations (those observations in our data that contain no missing data). We can do this a few different ways.
# data frame with missing values
df <- data.frame(col1 = c(1:3, NA),
col2 = c("this", NA,"is", "text"),
col3 = c(TRUE, FALSE, TRUE, TRUE),
col4 = c(2.5, 4.2, 3.2, NA))
df## col1 col2 col3 col4
## 1 1 this TRUE 2.5
## 2 2 <NA> FALSE 4.2
## 3 3 is TRUE 3.2
## 4 NA text TRUE NAFirst, to find complete cases we can leverage the
complete.cases() function which returns a logical vector
identifying rows which are complete cases. So, in the following case
rows 1 and 3 are complete cases. We can use this information to subset
our data frame which will return the rows which
complete.cases() found to be TRUE.
complete.cases(df)## [1] TRUE FALSE TRUE FALSE# subset data frame with complete.cases to get only complete cases
df[complete.cases(df), ]## col1 col2 col3 col4
## 1 1 this TRUE 2.5
## 3 3 is TRUE 3.2# or subset with `!` operator to get incomplete cases
df[!complete.cases(df), ]## col1 col2 col3 col4
## 2 2 <NA> FALSE 4.2
## 4 NA text TRUE NAA shorthand alternative approach is to simply use
na.omit() to omit all rows containing missing values.
# or use na.omit() to get same as above
na.omit(df)## col1 col2 col3 col4
## 1 1 this TRUE 2.5
## 3 3 is TRUE 3.2However, it seems like a waste to omit the information in all the
other fields just because one field value is missing. Therefore, data
analysts should carefully approach to excluding missing values
especially when the amount of missing data is very large.
Another recommended approach is to replace the missing value with a value substituted according to various criteria. These approaches will be given in the next section.
Basic Missing Value Imputation Techniques
Imputation is the process of estimating or deriving values for fields
where data is missing. There is a vast body of literature on imputation
methods and it goes beyond the scope of this course to discuss all of
them. In this section I will provide basic missing value imputation
techniques only.
Replace the missing value(s) with some constant, specified by the analyst
In some cases, a missing value can be determined because the observed
values combined with their constraints force a unique solution. As an
example, consider the following data frame listing the costs for
staff, cleaning, housing and the
total total for three months.
df <- data.frame(month = c(1:3),
staff = c(15000 , 20000, 23000),
cleaning = c(100, NA, 500),
housing = c(300, 200, NA),
total = c(NA, 20500, 24000))
df## month staff cleaning housing total
## 1 1 15000 100 300 NA
## 2 2 20000 NA 200 20500
## 3 3 23000 500 NA 24000Now, assume that we have the following rules for the calculation of
total cost: staff + cleaning +
housing = total and all costs > 0.
Therefore, if one of the variables is missing, we can clearly derive the
missing values by solving the rule. For this example, first month’s
total cost can be found as 15000 + 100 + 300 = 15400. Other missing
values can be found in a similar way.
The deductive and validate packages have a
number of functions available that can impute (and correct) the values
according to the given rules automatically for a given data frame.
#install.packages("deductive")
#install.packages("validate")
library(deductive)
library(validate)# Define the rules as a validator expression
Rules <- validator( staff + cleaning + housing == total,
staff >= 0,
housing >= 0,
cleaning >= 0)
# Use impute_lr function
imputed_df <- impute_lr(df,Rules)
imputed_df## month staff cleaning housing total
## 1 1 15000 100 300 15400
## 2 2 20000 300 200 20500
## 3 3 23000 500 500 24000The deducorrect package together with
validate provide a collection of powerful methods for
automated data cleaning and imputing. For more information on these
packages please refer to “Correction of Obvious Inconsistencies and
Errors” section of the module notes and the deducorrect
package manual and validate
package manual.
Replace the missing value(s) with the mean, median or mode
Replacing the missing value with the mean, median (for numerical
variables) or the mode (for categorical variables) is a crude way of
treating missing values. The Hmisc package has a convenient
wrapper function allowing you to specify what function is used to
compute imputed values from the non-missing.
Consider the following data frame with missing values:
x <- data.frame( no = c(1:6),
x1 = c(15000 , 20000, 23000, NA, 18000, 21000),
x2 = c(4, NA, 4, 5, 7, 8),
x3 = factor(c(NA, "False", "False", "False", "True", "True")))
x## no x1 x2 x3
## 1 1 15000 4 <NA>
## 2 2 20000 NA False
## 3 3 23000 4 False
## 4 4 NA 5 False
## 5 5 18000 7 True
## 6 6 21000 8 TrueFor this data frame, imputation of the mean, median and mode can be
done using Hmisc package as follows:
#install.packages("Hmisc")library(Hmisc)
# mean imputation (for numerical variables)
x1 <- impute(x$x1, fun = mean)
x1## 1 2 3 4 5 6
## 15000 20000 23000 19400* 18000 21000# median imputation (for numerical variables)
x2 <- impute(x$x2, fun = median)
x2## 1 2 3 4 5 6
## 4 5* 4 5 7 8# mode imputation (for categorical/factor variables)
x3 <- impute(x$x3, fun= mode)
x3## 1 2 3 4 5 6
## False* False False False True TrueA nice feature of the impute function is that the
resulting vector remembers what values were imputed. This information
may be requested with is.imputed function as in the example
below.
# check which values are imputed
is.imputed(x1)## [1] FALSE FALSE FALSE TRUE FALSE FALSEis.imputed(x2)## [1] FALSE TRUE FALSE FALSE FALSE FALSEis.imputed(x3)## [1] TRUE FALSE FALSE FALSE FALSE FALSEMore Complex Approaches to Missing Value Imputation
Another strategy is to use predictive models to impute the missing
data. There are many different predictive models and algorithms to
predict and impute the missing values. Regression
analysis, multiple imputation methods, random forests, k nearest neighbours, last observation
carried forward / next observation carried backward, etc. are only some
of these techniques. In R, there are many different packages (e.g.,
mice, missForest, impute etc.)
that can be used to predict and impute the missing data.
For the detailed information on the missing value imputation please refer to the “Statistical analysis with missing data (Little and Rubin (2014))” for the theory behind the missing value mechanism and analysis. For multiple imputation techniques and case studies using R, please refer to “Flexible imputation of missing data (Van Buuren (2012))”.
Special values
In addition to missing values, there are a few special values that
are used in R. These are -Inf, Inf and
NaN.
If a computation results in a number that is too big, R will return
Inf (meaning positive infinity) for a positive number and
-Inf for a negative number (meaning negative infinity).
Here are some examples:
3 ^ 1024## [1] Inf -3 ^ 1024## [1] -InfThis is also the value returned when you divide by 0:
12 / 0## [1] InfSometimes, a computation will produce a result that makes little
sense. In these cases, R will often return NaN (meaning
“not a number”):
Inf - Inf## [1] NaN0/0## [1] NaNIdentifying Special Values
Calculations involving special values often result in special values,
thus it is important to handle special values prior to analysis. The
is.finite, is.infinite, or is.nan
functions will generate logical values (TRUE or FALSE) and they can be
used to identify the special values in a data set.
# create a vector with special values
m <- c( 2, 0/0, NA, 1/0, -Inf, Inf, (Inf*2) )
m## [1] 2 NaN NA Inf -Inf Inf Inf# check finite values
is.finite(m)## [1] TRUE FALSE FALSE FALSE FALSE FALSE FALSE# check infinite (-inf or +inf) values
is.infinite(m)## [1] FALSE FALSE FALSE TRUE TRUE TRUE TRUE# check not a number (NaN) values
is.nan(m)## [1] FALSE TRUE FALSE FALSE FALSE FALSE FALSE# create a data frame containing special values
df <- data.frame(col1 = c( 2, 0/0, NA, 1/0, -Inf, Inf),
col2 = c( NA, Inf/0, 2/0, NaN, -Inf, 4))
df## col1 col2
## 1 2 NA
## 2 NaN Inf
## 3 NA Inf
## 4 Inf NaN
## 5 -Inf -Inf
## 6 Inf 4# check whether dataframe has infinite (-inf or +inf) values
is.infinite(df)
# Error in is.infinite(df) : default method not implemented for type 'list'These functions accept vectorial input, therefore you will receive an
error when you try to use it with a data frame. In such cases, we can
use apply family functions.
Remark: Useful apply family functions
The apply family functions will apply a specified
function to a given data object (e.g. vectors, lists, matrices, data
frames). Most common forms of apply functions are:
apply()for matrices and data frameslapply()for lists (output as list)sapply()for lists (output simplified)tapply()for vectors- … many others
There is a very useful and comprehensive
tutorial on apply family functions in DataCamp. Please
read this tutorial for more information on the usage of
apply family. You can also use swirl() to
practice apply family functions. In swirl(), “R
Programming Course” Lesson 10 and 11 cover the apply family
functions.
Among those apply functions, sapply and
lapply functions can apply any function to a list. Remember
from Module 3 that data frames possess the characteristics of both lists
and matrices. Therefore, we can use sapply or
lappy for data frames.
Now for the previous example, let’s use sapply
function:
df## col1 col2
## 1 2 NA
## 2 NaN Inf
## 3 NA Inf
## 4 Inf NaN
## 5 -Inf -Inf
## 6 Inf 4# check whether dataframe has infinite (-inf or +inf) values using sapply
sapply(df, is.infinite)## col1 col2
## [1,] FALSE FALSE
## [2,] FALSE TRUE
## [3,] FALSE TRUE
## [4,] TRUE FALSE
## [5,] TRUE TRUE
## [6,] TRUE FALSEBy using sapply we could see the infinite values in the
data frame. Now remember that is.infinite function doesn’t
check for NaN numbers. Therefore, we also need to check the
data frame for NaN values. In order to do that, we can
write a simple function to check every numerical column in a data frame
for infinite values or NaN’s.
# data frame
df## col1 col2
## 1 2 NA
## 2 NaN Inf
## 3 NA Inf
## 4 Inf NaN
## 5 -Inf -Inf
## 6 Inf 4# Check every numerical column whether they have infinite or NaN values using a function called is.special
is.special <- function(x){
if (is.numeric(x)) (is.infinite(x) | is.nan(x))
}
# apply this function to the data frame.
sapply(df, is.special)## col1 col2
## [1,] FALSE FALSE
## [2,] TRUE TRUE
## [3,] FALSE TRUE
## [4,] TRUE TRUE
## [5,] TRUE TRUE
## [6,] TRUE FALSEHere, the is.special function is applied to each column
of df using sapply. is.special
checks the data frame for numerical special values if the type is
numeric. Using a similar approach you can also check for special values
or NA’s at the same time using:
# Check every numerical column whether they have infinite or NaN or NA values using a function called is.specialorNA
is.specialorNA <- function(x){
if (is.numeric(x)) (is.infinite(x) | is.nan(x) | is.na(x))
}
# apply this function to the data frame.
sapply(df, is.specialorNA)## col1 col2
## [1,] FALSE TRUE
## [2,] TRUE TRUE
## [3,] TRUE TRUE
## [4,] TRUE TRUE
## [5,] TRUE TRUE
## [6,] TRUE FALSEEspecially for big datasets, the output of is.xxxx
functions would be very long as it returns TRUE or
FALSE values for each row and column in the data frame. To
avoid such non-informative and long output you can use
sum() function together with is.xxxx functions
and apply it to a data frame using lapply or
sapply. However, you cannot use (nested) functions inside
apply family functions. One solution to this is to write a
user defined function inside apply function.
Here is a good example on population_NA data
frame:
# a dataset with some NA values
population_NA## Region X.2013 X.2014 X.2015 X.2016
## 1 ISL 3.21 3.25 3.28 3.32
## 2 CAN 3.87 3.91 3.94 3.99
## 3 RUS 7.83 7.85 7.87 <NA>
## 4 COL 41.27 41.74 NA <NA>
## 5 ZAF 43.53 44.22 NA <NA>
## 6 LTU 47.42 46.96 46.63 46.11
## 7 MEX 60.43 61.10 61.76 62.41
## 8 IND 394.85 NA NA <NA>
## 9 NLD 497.64 499.59 501.68 504.01
## 10 KOR 504.92 506.97 508.91 510.77# let's check whether the data frame has NA values using sapply
sapply(population_NA, is.na)## Region X.2013 X.2014 X.2015 X.2016
## [1,] FALSE FALSE FALSE FALSE FALSE
## [2,] FALSE FALSE FALSE FALSE FALSE
## [3,] FALSE FALSE FALSE FALSE TRUE
## [4,] FALSE FALSE FALSE TRUE TRUE
## [5,] FALSE FALSE FALSE TRUE TRUE
## [6,] FALSE FALSE FALSE FALSE FALSE
## [7,] FALSE FALSE FALSE FALSE FALSE
## [8,] FALSE FALSE TRUE TRUE TRUE
## [9,] FALSE FALSE FALSE FALSE FALSE
## [10,] FALSE FALSE FALSE FALSE FALSEWe can avoid long output and calculate the total number of missing
values for each column using sum() and is.na()
functions.
# Note that the code given below would throw an error
sapply(population_NA, sum(is.na()))
# Error in is.na() : 0 arguments passed to 'is.na' which requires 1Instead you can write your own function inside sapply()
and calculate the total missing values for each column:
sapply(population_NA, function(x) sum( is.na(x) ))## Region X.2013 X.2014 X.2015 X.2016
## 0 0 1 3 4Optional Reading: Checking for Obvious Inconsistencies or Errors
An obvious inconsistency occurs when a data record contains a value
or combination of values that cannot correspond to a real-world
situation. For example, a person’s age cannot be negative, a man cannot
be pregnant, and an under-aged person cannot possess a drivers’ license.
Such knowledge can be expressed as rules or constraints. In data
preprocessing literature these rules are referred to as edit
rules or edits, in short. Checking for obvious
inconsistencies can be done straightforwardly in R using logical
indices.
For example, to check which elements of x obey the rule:
“x must be non-negative” one can simply use the
following.
# create a vector called x
x <- c( 0, -2, 1, 5)
# check the non-negative elements
x_nonnegative <- (x >= 0)
x_nonnegative## [1] TRUE FALSE TRUE TRUEHowever, as the number of variables increases, the number of rules
may increase, and it may be a good idea to manage the rules separate
from the data. For such cases, the editrules package allows
us to define rules on categorical, numerical or mixed-type data sets
which each record must obey. Furthermore, editrules can
check which rules are obeyed or not and allows one to find the minimal
set of variables to adapt so that all rules can be obeyed. This package
also implements several basic rule operations allowing users to test
rule sets for contradictions and certain redundancies.
To illustrate I will use a small data set (datawitherrors.csv) given below:
datawitherrors <- read.csv("data/datawitherrors.csv")
datawitherrors## no age agegroup height status yearsmarried
## 1 1 21 adult 178 single -1
## 2 2 2 child 147 married 0
## 3 3 18 adult 167 married 20
## 4 4 221 elderly 154 widowed 2
## 5 5 34 child -174 married 3As you noticed, there are many inconsistencies/errors in this small
data set (i.e., age = 221, height = -174, years married = -1, etc.) . To
begin with a simple case, let’s define a restriction on the age variable
using editset functions. In order to use
editset functions, we need to install and load the
editrules package.
#install.packages("editrules")
library(editrules)In the first rule, we will define the restriction on the age variable
as $ 0 age $ using editset function.
(Rule1 <- editset(c("age >= 0", "age <= 150")))##
## Edit set:
## num1 : 0 <= age
## num2 : age <= 150The editset function parses the textual rules and stores
them in an editset object. Each rule is assigned a name according to its
type (numeric, categorical, or mixed) and a number. The data set can be
checked against these rules using the violatedEdits
function.
violatedEdits(Rule1, datawitherrors)## edit
## record num1 num2
## 1 FALSE FALSE
## 2 FALSE FALSE
## 3 FALSE FALSE
## 4 FALSE TRUE
## 5 FALSE FALSEviolatedEdits returns a logical array indicating for
each row of the data, which rules are violated. From the output, it can
be understood that the 4th record violates the second rule (age <=
150).
One can also read rules, directly from a text file using the
editfile function. As an example, consider the contents of
the following text file (also available here):
1 # numerical rules
2 age >= 0
3 height > 0
4 age <= 150
5 age > yearsmarried
6
7 # categorical rules
8 status %in% c(“married”,“single”,“widowed”)
9 agegroup %in% c(“child”,“adult”,“elderly”)
10 if ( status == “married” ) agegroup %in% c(“adult”,“elderly”)
11
12 # mixed rules
13 if ( status %in% c(“married”,“widowed”)) age - yearsmarried >=
17
14 if ( age < 18 ) agegroup == “child”
15 if ( age >= 18 && age <65 ) agegroup == “adult”
16 if ( age >= 65 ) agegroup == “elderly”
These rules are numerical, categorical and mixed (both data types).
Comments are written behind the usual # character. The rule
set can be read using editfile function as follows:
Rules <- editfile("data/editrules.txt", type = "all")
Rules##
## Data model:
## dat6 : agegroup %in% c('adult', 'child', 'elderly')
## dat7 : status %in% c('married', 'single', 'widowed')
##
## Edit set:
## num1 : 0 <= age
## num2 : 0 < height
## num3 : age <= 150
## num4 : yearsmarried < age
## cat5 : if( agegroup == 'child' ) status != 'married'
## mix6 : if( age < yearsmarried + 17 ) !( status %in% c('married', 'widowed') )
## mix7 : if( age < 18 ) !( agegroup %in% c('adult', 'elderly') )
## mix8 : if( 18 <= age & age < 65 ) !( agegroup %in% c('child', 'elderly') )
## mix9 : if( 65 <= age ) !( agegroup %in% c('adult', 'child') )violatedEdits(Rules, datawitherrors)## edit
## record num1 num2 num3 num4 dat6 dat7 cat5 mix6 mix7 mix8 mix9
## 1 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## 2 FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE
## 3 FALSE FALSE FALSE TRUE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
## 4 FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## 5 FALSE TRUE FALSE FALSE FALSE FALSE TRUE FALSE FALSE TRUE FALSEAs the number of rules grows, looking at the full array produced by
violatedEdits becomes complicated. For this reason,
editrules offers methods to summarise or visualise the
result as follows:
Violated <- violatedEdits(Rules, datawitherrors)
# summary of violated rules
summary(Violated)## Edit violations, 5 observations, 0 completely missing (0%):
##
## editname freq rel
## cat5 2 40%
## mix6 2 40%
## num2 1 20%
## num3 1 20%
## num4 1 20%
## mix8 1 20%
##
## Edit violations per record:
##
## errors freq rel
## 0 1 20%
## 1 1 20%
## 2 2 40%
## 3 1 20%# plot of violated rules
plot(Violated)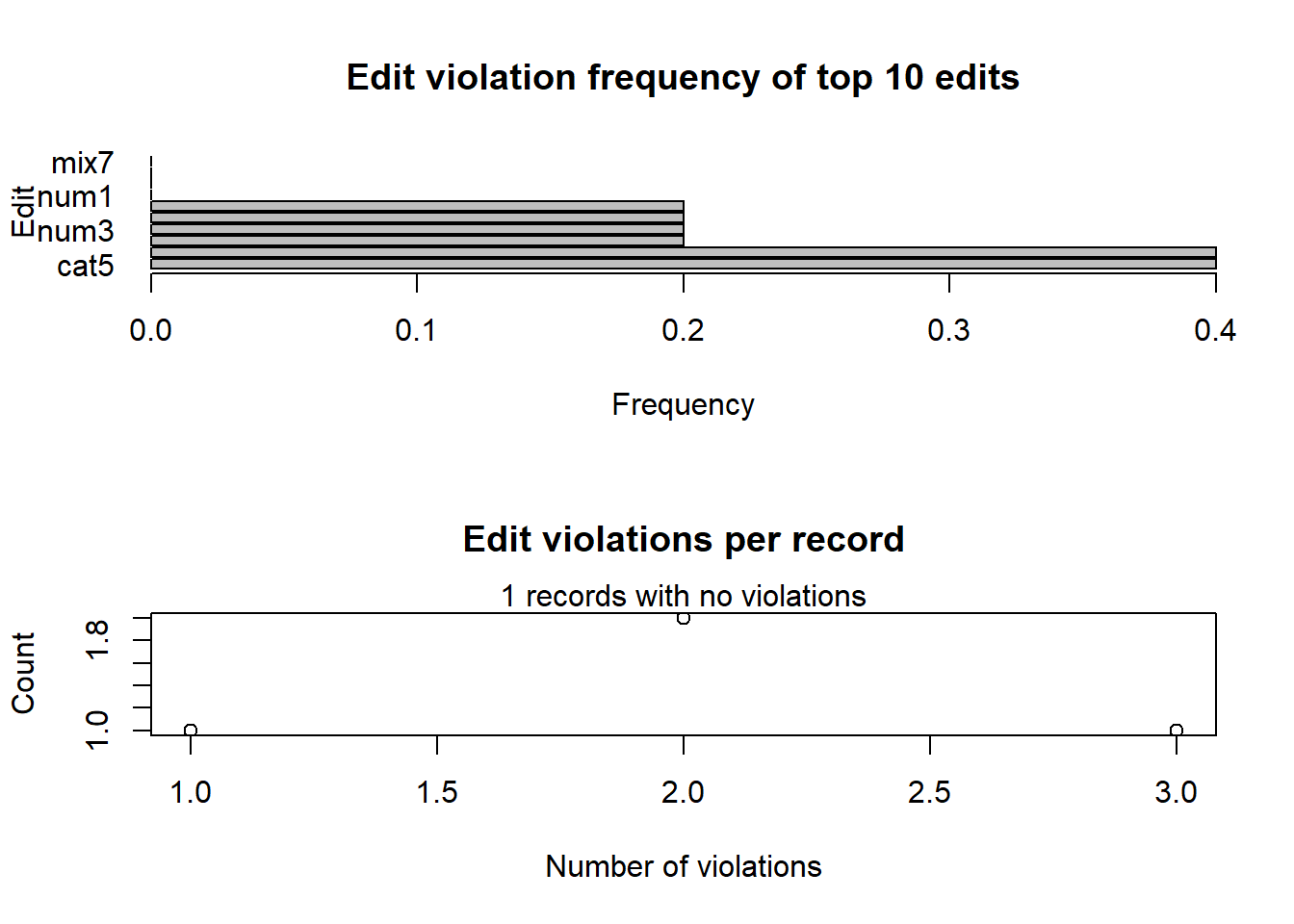
Using the functions available in editrules package,
users can detect the obvious errors and/or inconsistencies in the data
set and define edit rules to identify the inconsistent records.
Moreover, analysts may need to correct the obvious errors and/or
inconsistencies in a data set. In the next section, I will introduce the
deductive package functions to correct the obvious errors
and inconsistencies.
Optional Reading: Correction of Obvious Inconsistencies or Errors
When the data you are analysing is generated by people rather than machines or measurement devices, certain typical human-generated errors are likely to occur. Given that data must obey certain edit rules, the occurrence of such errors can sometimes be detected from raw data with (almost) certainty. Examples of errors that can be detected are typing errors in numbers, rounding errors in numbers, and sign errors.
The deducorrect package has several functions available
that can correct such errors. Consider the following data frame (datawitherrors2.csv):
datawitherrors2 <- read.csv("data/datawitherrors2.csv")
datawitherrors2## no height unit
## 1 1 178.00 cm
## 2 2 1.47 m
## 3 3 70.00 inch
## 4 4 154.00 cm
## 5 5 5.92 ftThe task here is to standardise the lengths and express all of them
in meters. The deducorrect package can correct this
inconsistency using correctionRules function. For example,
to perform the above task, one first specifies a file with correction
rules as follows (also available here).
1 # convert centimeters
2 if ( unit == “cm” ){
3 height <- height/100
4 }
5 # convert inches
6 if (unit == “inch” ){
7 height <- height/39.37
8 }
9 # convert feet
10 if (unit == “ft” ){
11 height <- height/3.28
12 }
13 # set all units to meter
14 unit <- “m”
With correctionRules we can read these rules from the
txt file using .file argument.
#install.packages("deducorrect")
library(deducorrect)# read rules from txt file using validate
Rules2 <- correctionRules("data/editrules2.txt")
Rules2## Object of class 'correctionRules'
## ## 1-------
## if (unit == "cm") height <- height/100
## ## 2-------
## if (unit == "inch") height <- height/39.37
## ## 3-------
## if (unit == "ft") height <- height/3.28
## ## 4-------
## unit <- "m"Now, we can apply them to the data frame and obtain a log of all
actual changes as follows:
cor <- correctWithRules(Rules2, datawitherrors2)
cor## $corrected
## no height unit
## 1 1 1.780000 m
## 2 2 1.470000 m
## 3 3 1.778004 m
## 4 4 1.540000 m
## 5 5 1.804878 m
##
## $corrections
## row variable old new how
## 1 1 height 178 1.78 if (unit == "cm") height <- height/100
## 2 1 unit cm m unit <- "m"
## 3 3 height 70 1.778004 if (unit == "inch") height <- height/39.37
## 4 3 unit inch m unit <- "m"
## 5 4 height 154 1.54 if (unit == "cm") height <- height/100
## 6 4 unit cm m unit <- "m"
## 7 5 height 5.92 1.804878 if (unit == "ft") height <- height/3.28
## 8 5 unit ft m unit <- "m"The returned value, cor$corrected will give a list
containing the corrected data as follows:
cor$corrected## no height unit
## 1 1 1.780000 m
## 2 2 1.470000 m
## 3 3 1.778004 m
## 4 4 1.540000 m
## 5 5 1.804878 mAdditional Resources and Further Reading
As mentioned before, the missing value analysis and the missing value imputation are broader concepts that would be a standalone topic of another course. Interested readers may refer to the “Statistical analysis with missing data (Little and Rubin (2014))” and “Flexible imputation of missing data (Van Buuren (2012))” for the theory behind the missing value mechanism and analysis.
There are many good R tutorials for handling missing data using R.
“Data
Science Live Book” are only two of them. Moreover, the missForest
and mice
packages’ manuals provide detailed information on the missing value
imputation using random forest algorithm and multiple imputation
techniques, respectively.
For checking and correcting errors and inconsistencies in the data,
users can refer to the deducorrect
, deductive
and validate
packages’ manuals and “An
introduction to data cleaning with R (De Jonge
and Loo (2013))” discussion paper.